


Efficient and scalable Iceberg I/O is crucial for Python data workloads using Iceberg, yet scaling Python to handle large datasets has traditionally involved significant trade-offs. While PyIceberg offers a Pythonic experience, it struggles to scale beyond a single node. Conversely, Spark provides the necessary scalability but lacks native Python ergonomics. More recently, tools like Daft have tried to bridge this gap, but they introduce new APIs instead of Pandas and their scaling and performance capabilities are not clear yet.
The Bodo DataFrame library bridges this gap by acting as a drop-in replacement for Pandas, seamlessly scaling native Python code across multiple cores and nodes using high-performance computing (HPC) techniques. This approach eliminates the need for JVM dependencies or changes in syntax, ensuring an efficient, ergonomic solution.
In this post, we benchmark Iceberg I/O performance across several Python-compatible engines—Bodo, Spark, PyIceberg, and Daft—focusing on reading and writing large Iceberg tables stored in Amazon S3 Tables. Our findings demonstrate that Bodo outperforms Spark by up to 3x, while PyIceberg and Daft were unable to complete the benchmark.
Bodo is an open-source, high-performance DataFrame library for Python that is a drop-in replacement for Pandas. Bodo simplifies accelerating and scaling Python workloads from laptops to clusters without code rewrites. Under the hood, Bodo relies on MPI-based high-performance computing (HPC) technology and an innovative auto-parallelizing just-in-time (JIT) compiler—making it both easier to use and many times faster than tools like Spark.
We evaluated the performance of copying a large Iceberg table using four tools:
The dataset was a TPC-H SF1000 Iceberg table (1.5 billion rows, 9 columns, ~12,000 files) stored in Amazon S3 Tables.
We tested both single-node and four-node configurations using c6i.32xlarge instances (128 vCPUs, 256 GB RAM, 50 Gbps network bandwidth). The benchmark used a simple table copy operation to evaluate IO performance end-to-end. We used the default configurations of all tools.
Code for our benchmark can be found in our Github repository.

PyIceberg and Daft were unable to complete the workload. PyIceberg lacks multi-node support and timed out after 12+ hours on a single node. Daft/Ray ran out of memory on both 1-node and 4-node configurations, likely because of the lack of streaming when using the Ray executor. By contrast, Bodo completed the job in under 45 minutes on a single node and under 12 minutes on a 4-node cluster, with no special tuning or JVM dependency. We will follow up with the Daft team to understand the potential current issues of Daft for this workload and if there is some configuration that could help.
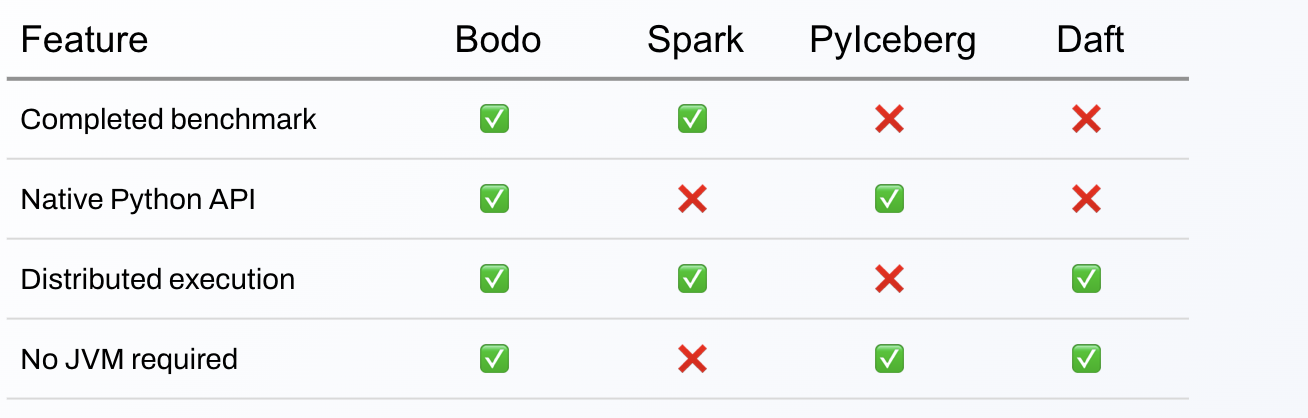
This table summarizes the features of the tested engines for this use case.
Bodo’s advantage in this benchmark is due to its fundamentally different architecture and design:
The Bodo code for read and write is compatible with Pandas but others require new APIs. The read and write in Bodo is simply:
orders_table = pd.read_iceberg("sf1000.orders", location=s3tables_arn)
orders_table.to_iceberg("sf1000.orders_copy_bodo", location=s3tables_arn)
Python data processing can be both easy and scalable using a DataFrame library like Bodo that is designed with native Python and efficient parallel architecture principles. If you are interested in Bodo: